Search EzBioCloud Database
Featured services
Other services
*Do you need NGS service? Please contact us at bs.ngs@cj.net
Recent blogs
-
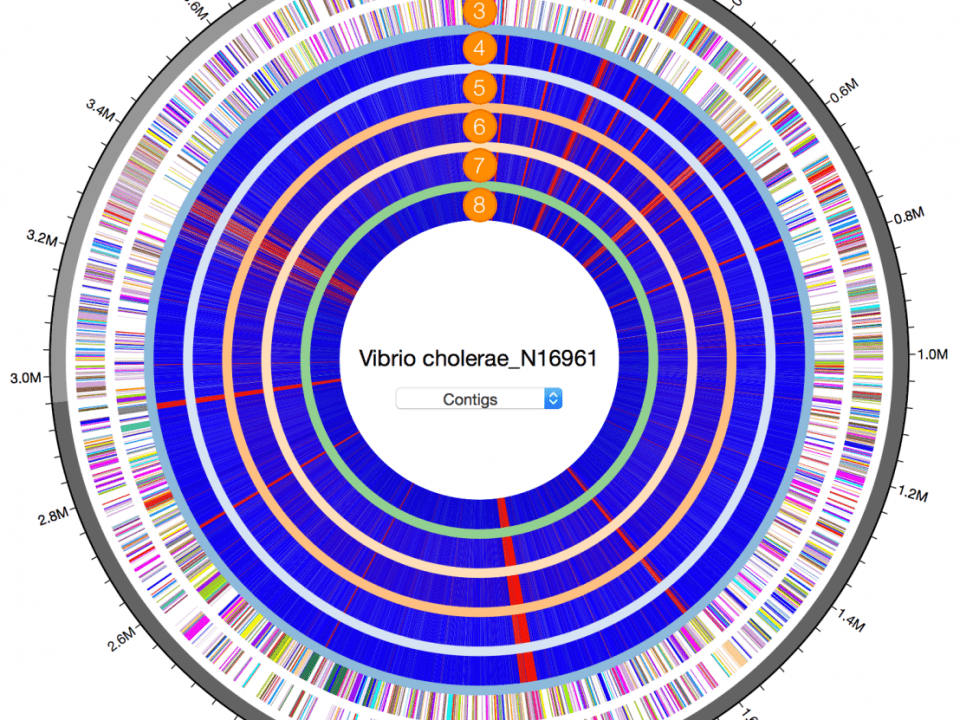
Genome-based Identification for Improving Reference Databases
Misidentified or incompletely identified bacterial genome sequences appear frequently in public reference databases. These databases can be significantly improved by genome-based identification against an up-to-date, systematically curated reference database that covers as many as species. -
Bacterial Nomenclature 101 and how to describe new species
Bacterial nomenclature and naming are regulated by the International Code of Nomenclature of Prokaryotes (shortly the Code), but not the actual species is. For example, the name Escherichia coli is regulated by the code, but not the species itself.
Times Cited
Registered Users
Identifications provided so far


















 This browser is not supported.
This browser is not supported.
